By comparing this score to the distribution of scores in globular and coiled-coil proteins, the program then calculates the probability that the sequence will adopt a coiled-coil conformation. Solvent accessibility is predicted by a neural network method rating at a correlation coefficient correlation between experimentally observed and predicted relative solvent accessibility of 0. A comparison with a statistical method previously described and tested on the same data base shows that the neural network-based predictor is performing with the highest efficiency. PredictProtein is the acronym for all prediction programs run. Technical tricks recurrent connections, shared weights etc. Transmembrane helices PHDhtm; example for output Transmembrane helices in integral membrane proteins are predicted by a system of neural networks. The output of the neural network codes for 10 states of relative accessibility. 
| Uploader: | Galabar |
| Date Added: | 17 April 2013 |
| File Size: | 29.17 Mb |
| Operating Systems: | Windows NT/2000/XP/2003/2003/7/8/10 MacOS 10/X |
| Downloads: | 66392 |
| Price: | Free* [*Free Regsitration Required] |
Low-complexity segments pudsec by the algorithm represent "simple sequences" or "compositionally-biased regions". For more information, you may have a look at the preliminary preprint. The principle idea is to detect similar motifs of secondary structure and accessibility between a sequence of unknown structure and a known fold.
Prediction accuracy is higher than average for eukaryotic proteins and lower than average for prokaryotes.
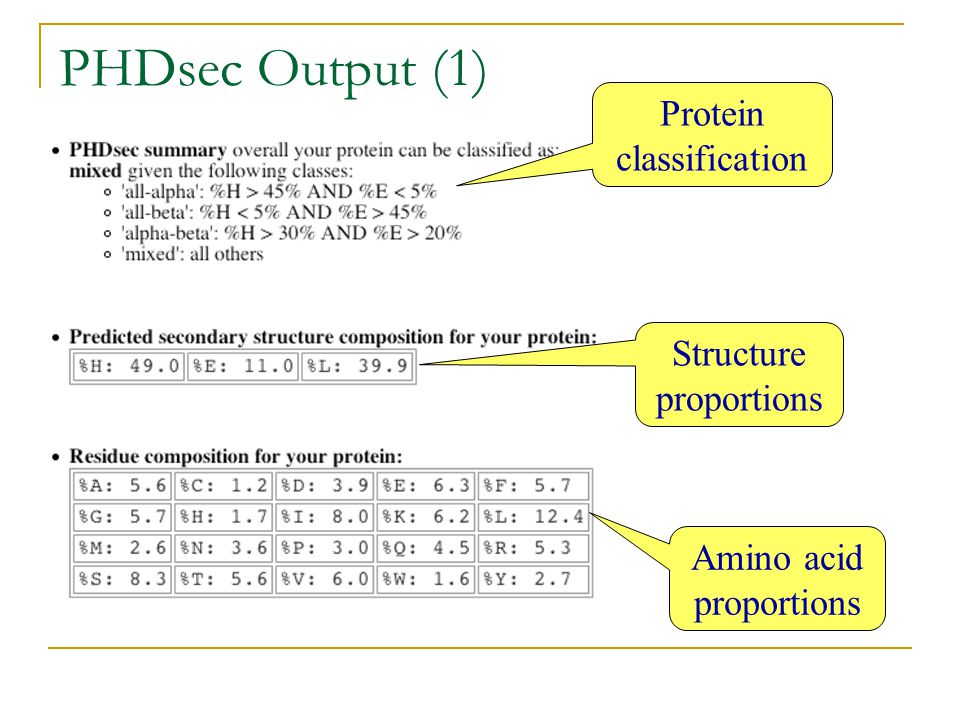
Accuracy of PHDsec
Nucleic Acids Res, 28, Bioinformatics, 15, Methods in Enzymology, phcsec Role of evolutionary information in predicting the disulfide-bonding state of cysteine in proteins.
Protein fold recognition by prediction-based threading. J Molecular Biol, PHDsec predictions have three main features: Applications and statistics for multiple high-scoring segments in molecular sequences.
The number of false positives, i. It has been devised to assist with the analysis of the domain arrangement of proteins.
PHDsec: secondary structure prediction
PrecitNLS finds experimentally known nuclear localisation in your protein. Transmembrane helices in integral membrane proteins are predicted by a system of neural networks. J Mol Biol2: The following description is from the original ProDom site which supplies a rather useful graphical interface to the ProDom database: Functional sequence motifs ProSite; example for output The following description is from the original ProSite site: Improved version of PHD: Low-complexity regions typically correspond to 'simple sequences' or 'compositionally-biased' regions.
Bioinformatics, 14, It consists of a database of biologically significant sites, patterns and profiles that help to reliably identify to which known family of protein if any a new sequence belongs. Pdsec is a program that compares a sequence to a database of known parallel two-stranded coiled-coils and derives a similarity score.
PP: details for methods used
The shortcoming of the network system is that often too long helices are predicted. Comput Appl Biosci, 11 6: This program can be run through the PredictProtein service. The shortcoming of the network system is that often too long helices are predicted. Combinaison de classifieurs statistiques, Application a la prediction de structure secondaire des proteines.
These are cut by an empirical filter. In sweep 2, after all sequences with significant homology have been picked from the BLASTP output, the profile is recompiled, and the dynamic programming algorithm starts once again to align consecutively the sequences, this time using the conservation profile as derived after completion of sweep 1.
AAAI Press, The time it takes before your results are returned depends on the speed of your internet connection, the network speed on your new or refurbished computersthe length of your target sequence, and the complexity of the algorithm. Protein Sci, 5 The neural network prediction of transmembrane helices PHDhtm is refined by a dynamic programming-like algorithm. Protein Eng, 1 4:


Comments
Post a Comment